Coadd 1D Spectra
Overview
This document will describe how to combine the 1D spectra from multiple exposures of the same object.
Coadding 1D spectra must be done outside of the data reduction pipeline (run_pypeit); i.e. PypeIt will not coadd your spectra as part of the data reduction process.
The current defaults use the Optimal extraction and flux-calibrated data.
See below for the Current Coadd1D Data Model.
coadd1d File
The pypeit_coadd_1dspec script requires an input file to guide the process. The format of this type of Input File Format includes a Parameter Block (optional) and a Data Block (required).
We describe each in turn.
Parameter Block
This optional Parameter Block can set the name of the output file and/or modify default parameters for the script, see Coadd1DPar Keywords.
Here is an example:
# User-defined coadding parameters
[coadd1d]
coaddfile = 'J1217p3905_coadd.fits'
sensfuncfile = 'sensfunc.fits' # Required only for Echelle
The coaddfile parameter is optional, if it is not set
the output file will be placed in the same directory as the first spec1d
file named coadd1d_<target>_<instrument>_<start date>_<end date>.fits.
Note
If you are using an Echelle spectrograph
(e.g. keck_nires), you must specify a sensfuncfile generated by
Fluxing a standard star.
See Parameters below for common parameters modified for coadd1d.
Data Block
A Data Block is required to indicate the spectra
to be combined. It always begins and ends with coadd1d read and coadd1d end, respectively.
One should then include the path to the Spec1D Output files,
unless you intend to run the script within the same folder as the data.
Last, the data block provides a table of Spec1D Output files and the object name in each to be coadded. See Spec1D Output for a discussion of the naming.
Here is an example:
# Data block
coadd1d read
path /path/to/your/reduced/data/Science
filename | obj_id
spec1d_b27-J1217p3905_KASTb_2015May20T045733.560.fits | SPAT0176-SLIT0000-DET01
spec1d_b28-J1217p3905_KASTb_2015May20T051801.470.fits | SPAT0175-SLIT0000-DET01
coadd1d end
It is possible to provide only a single entry (it must be the first row)
in the obj_id column.
This will then be used for all of the Spec1D Output files input.
The list of object identifiers in a given Spec1D Output file can be output with the pypeit_show_1dspec script, e.g.:
pypeit_show_1dspec spec1d-filename.fits --list
These can also be recovered from the object info .txt files
in the Science/ folder (one per exposure).
pypeit_coadd_1dspec script
The primary script for 1D coadding is called pypeit_coadd_1dspec,
which requires a coadd1d File to guide the process.
The script usage can be displayed by calling the script with the
-h option:
$ pypeit_coadd_1dspec -h
usage: pypeit_coadd_1dspec [-h] [--debug] [--show] [--par_outfile PAR_OUTFILE]
[-v VERBOSITY]
coadd1d_file
Coadd 1D spectra produced by PypeIt
positional arguments:
coadd1d_file File to guide coadding process.
------------------------
MultiSlit
------------------------
For coadding Multislit spectra the file must have the
following format (see docs for further details including
the use of paths):
[coadd1d]
coaddfile='output_filename.fits' # Optional
coadd1d read
filename | obj_id
spec1dfile1 | objid1
spec1dfile2 | objid2
spec1dfile3 | objid3
...
coadd1d end
OR the coadd1d read/end block can look like
coadd1d read
filename | obj_id
spec1dfile1 | objid
spec1dfile2 |
spec1dfile3 |
...
coadd1d end
That is the coadd1d block must be a two column list of
spec1dfiles and objids, but you can specify only a
single objid for all spec1dfiles on the first line
Where:
spec1dfile: full path to a PypeIt spec1dfile
objid: the object identifier. To determine the objids
inspect the spec1d_*.txt files or run pypeit_show_1dspec
spec1dfile --list
------------------------
Echelle
------------------------
For coadding Echelle spectra the file must have the
following format (see docs for further details):
[coadd1d]
coaddfile='output_filename.fits' # Optional
coadd1d read
filename | obj_id | sensfile | setup_id
spec1dfile1 | objid1 | sensfile1 | setup_id1
spec1dfile2 | objid2 | sensfile2 | setup_id2
spec1dfile3 | objid3 | sensfile3 | setup_id3
...
coadd1d end
OR the coadd1d read/end block can look like
coadd1d read
filename | obj_id | sensfile | setup_id
spec1dfile1 | objid1 | sensfile | setup_id
spec1dfile2 | | |
spec1dfile3 | | |
...
coadd1d end
That is the coadd1d block is a four column list of
spec1dfiles, objids, sensitivity function files, and
setup_ids, but you can specify only a single objid,
sensfile, and setup_id for all spec1dfiles on the first
line
Here:
spec1dfile: full path to a PypeIt spec1dfile
objid: the object identifier (see details above)
sensfile: full path to a PypeIt sensitivity function
file for the echelle setup in question
setup_id: string identifier for the echelle setup in
question, i.e. 'VIS', 'NIR', or 'UVB'
If the coaddfile is not given the output file will be
placed in the same directory as the first spec1d file.
options:
-h, --help show this help message and exit
--debug show debug plots?
--show show QA during coadding process
--par_outfile PAR_OUTFILE
Output to save the parameters
-v VERBOSITY, --verbosity VERBOSITY
Verbosity level between 0 [none] and 2 [all]. Default:
1. Level 2 writes a log with filename
coadd_1dspec_YYYYMMDD-HHMM.log
run
An example execution looks like this:
pypeit_coadd_1dspec FRB190714_LRISr_coadd1d_file.txt --show
A substantial set of output are printed to the screen, and if successful the final spectrum is written to disk. See below for the Current Coadd1D Data Model.
The parameters that guide the coadd process are also written
to disk for your records. The default location is coadd1d.par.
You can choose another location with the --par_outfile
option (see below).
Command Line Options
–show
At the end of the process, this will launch a matplotlib window showing the stacked spectrum on the bottom. The top panel illustrates the number of pixels included in the stack.
–par_outfile
This file will hold a listing of the parameters used to run the coadd1d process.
Parameters
Here we describe a set of parameters frequently defined in the Parameter Block of the coadd1d File.
Fluxing
The default parameters assume your spectra have gone through Fluxing. If not you should set:
[coadd1d]
flux_value = False
If the data were fluxed, then the output flux spectrum will have units of
\([10^{-17} {\rm erg/s/cm^2/\mathrm{\mathring{A}}}]\).
Flux Scale
If your data has been fluxed, you may scale the coadded spectrum to a chosen value (typically a photometric measurement) in one of many filter curves.
To do so, you need to add the filter and magnitude
to the [coadd1d] block of the coadd1d file.
Here is an example:
[coadd1d]
coaddfile = 'J121555.09-130116.0_LRISr_A.fits'
filter = PS1-R
filter_mag = 20.85
filter_mask = 7187:7376
The call here will convolve the coadded spectrum with the PS1 r-band filter, and then scale the flux to give an AB magnitude of 20.85. Furthermore, the spectral wavelengths from \(7187 < \lambda < 7376 [\mathrm{\mathring{A}}]\) are masked in the analysis.
Filters
The list of available filters is provided in this file.
Scaling
Parameter |
Option |
Description |
|---|---|---|
scale_method |
default: auto |
scale the flux arrays based on the root mean square value (RMS) of the S/N^2 value for all spectra; if this RMS value is less than the minimum median scale value, no scaling is applied. If the RMS value is greater than the minimum but smaller than the maximum median scale value, the applied method is the median, as described below |
– |
hand |
scale the flux arrays using values specified by the user in the input parameter ‘hand_scale’. Must have one value per spectrum |
– |
median |
scale the flux arrays by the median flux value of each spectra |
Wave Method
You may want to specify the method used to construct the wavelength grid for
coadding your spectra. This is done by modifying wave_method in the
[coadd1d] block of your coadd1d file. The default method is linear,
which uses a fixed linear grid in wavelength. However, this may not be ideal
depending on your instrument set-up and observations.
Here is an example of some Keck LRISb data. After coadding the 1D spectra, you may see a noise pattern like this:
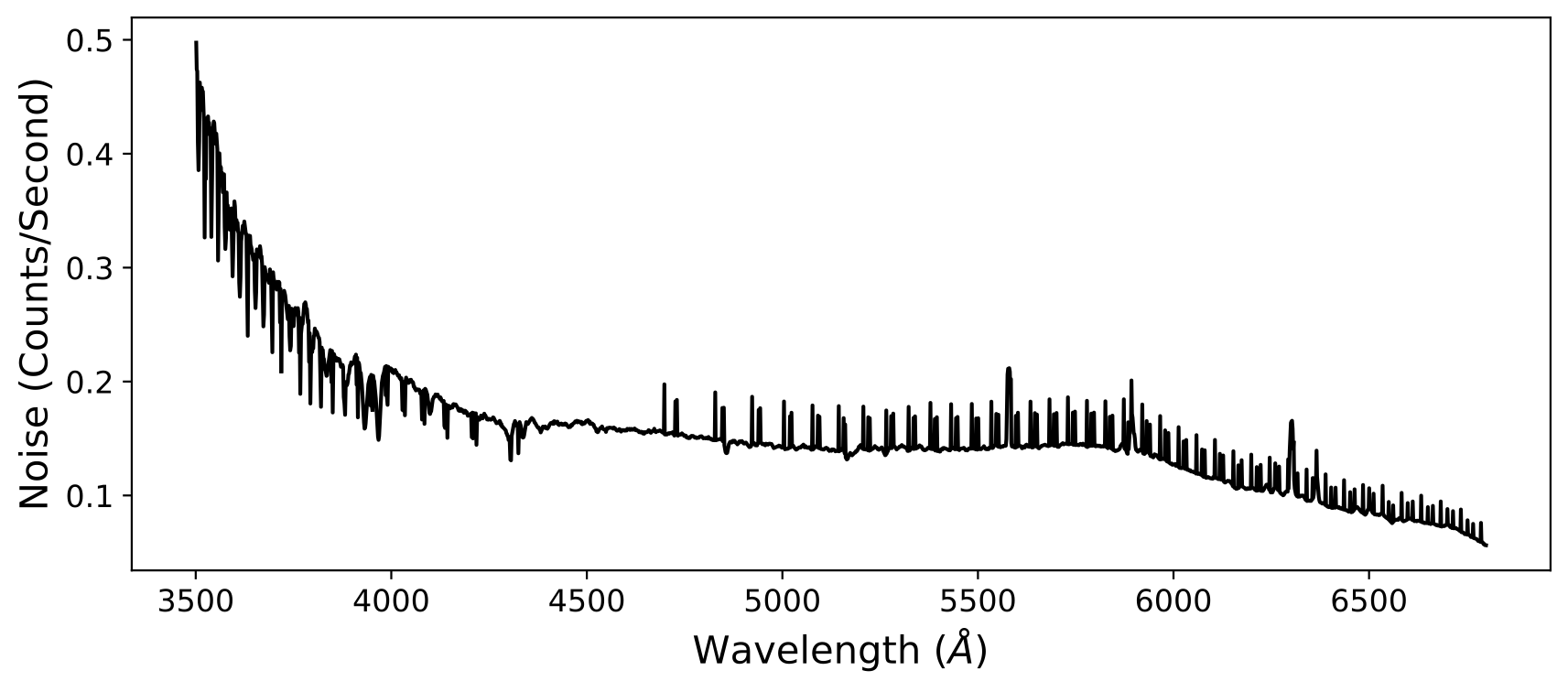
which, while still correct, may not be desirable. Note this figure was obtained
by plotting the quantity \(\frac{1}{\sqrt{\texttt{ivar}}}\) using the
ivar array in the second extension of the coadd1d file. This noise
pattern occurs because pypeit_coadd_1dspec does not interpolate the spectra,
in order to guarantee that neighboring pixels do not have correlated noise.
However, if your wavelength grid is such that multiple values land in one
re-binned pixel, this pattern will appear.
To avoid this noise pattern, you can modify the value of wave_method. For
example, if we use the iref option instead, the above example becomes:
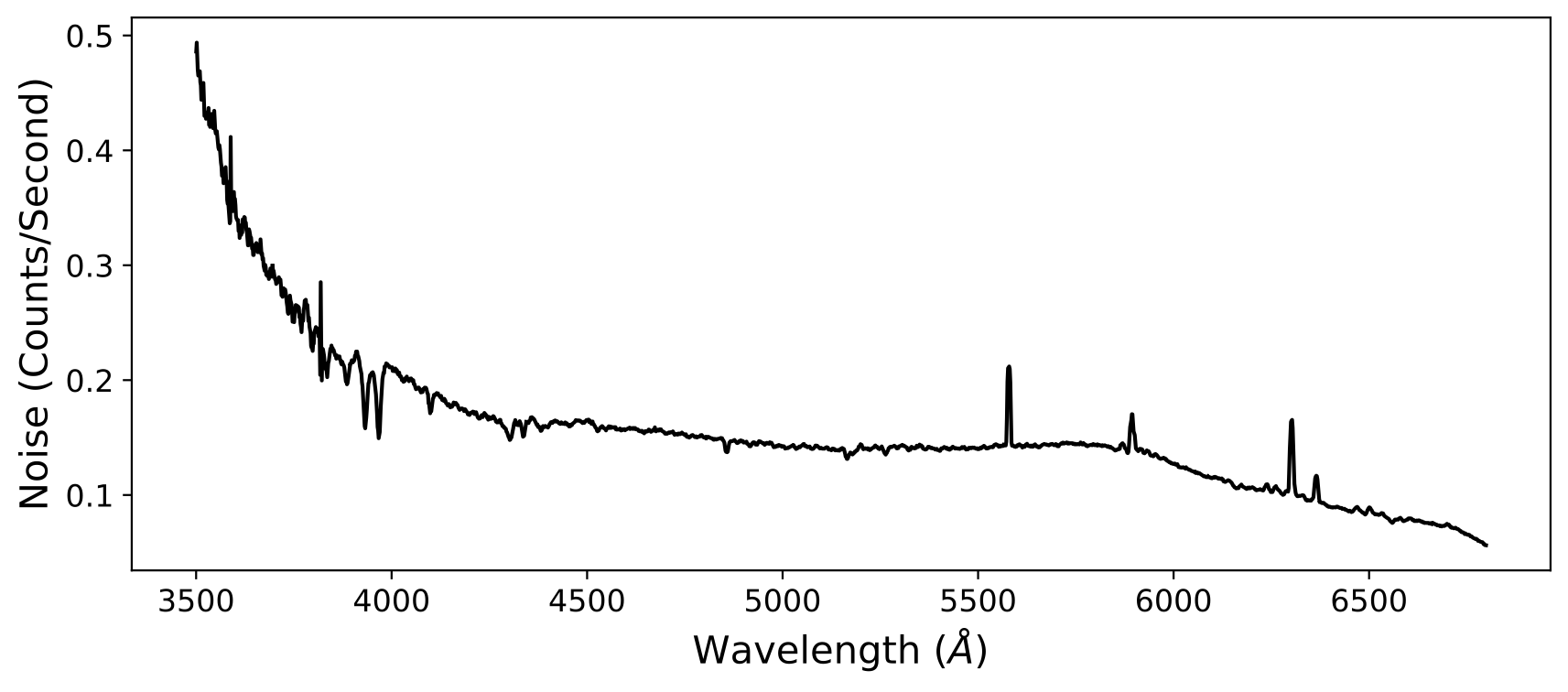
The iref option uses the grid of one of the spectra that you are coadding as
the wavelength grid. The reason why this works is because, in general, the
wavelength solution of a slit will be non-linear, so the pixel spacing will be
different between different wave_method options. For a linear grid, the
number of exposures contributing to a given pixel in the final grid is varying
because of how non-linear grids overlap with the linear grid. By using the
iref option or a linear grid with wider spacing, this effect can be reduced.
If your data contains exposures that aren’t dithered, the iref option will
coadd your exposures on the native wavelength grid, which will avoid the
fluctuations in the noise vector.
If your data contains exposures that are dithered a lot, iref will probably
give you similar noise fluctuations, so using a linear grid with wider spacing
may give you a better result. To change the spacing of the wavelength grid, you
can modify samp_fact in the [coadd1d] block of your coadd1d file. The
default value is 1.0. Setting samp_fact > 1.0 over-samples (finer grid),
while setting samp_fact < 1.0 under-samples (coarser grid).
Current Coadd1D Data Model
The result of the 1D coadding will be saved to a fits file. The file name can
be provided directly using the coadd1dfile parameter in the
coadd1d File or you can use the default. The default filename convention
is coadd1d_<target>_<instrument name>_<YYYYMMDD>.fits or
coadd1d_<target>_<instrument name>_<YYYYMMDD>-<YYYYMMDD>.fits, if the coadd
included more than one day’s worth of data. The default location of the file
will be along side the first spec1d file.
Currently instrument_name is
taken from the camera attribute of the relevant
Spectrograph class.
The format of the 1D coadd file follows the general
class OneSpec, such
that its file extensions are:
Version 1.0.2
HDU Name |
HDU Type |
Data Type |
Description |
|---|---|---|---|
|
… |
Empty data HDU. Contains basic header information. |
|
|
… |
Single spectrum data; see |
You view the spectrum using the lt_xspec script
(pypeit_show_1dspec will not work), which loads the data
and launches a GUI from the linetools package. e.g.:
lt_xspec J1217p3905_coadd.fits